Step-by-step Guidance to Run GUANACO
Command-line Usage
Basic command-line options:
options:
-h, --help Show this help message and exit
-c CONFIG, --config CONFIG
Name of configuration JSON file (relative to --data-dir)
(default: guanaco.json)
-d DATA_DIR, --data-dir DATA_DIR
Directory containing AnnData files referenced in config
(default: current directory)
-p PORT, --port PORT Port to run the Dash server on (default: 4399)
--host HOST Host to run the Dash server on (default: 0.0.0.0)
--debug Run server in debug mode (default: False)
--max-cells MAX_CELLS Maximum number of cells to load per dataset (default: 10000)
--backed-mode Enable backed mode for memory-efficient loading of
large datasets (default: False)
Tip
Quick start: If the config file and data files are in the current directory, you can simply run:
guanaco -c config.json
Config File Structure
The GUANACO configuration file is written in JSON format and has two main sections:
Studies
Each study is defined by a dictionary entry, where the key is the study name. A study can include either matrix-based data, track-based data, or both:
description (optional but recommended)
A short text description of the study. Displayed in the interface to help distinguish between datasets.
Matrix-based data
sc_data(required): Name of a.h5ador.h5mufile.markers(optional): List of marker genes for plots (heatmaps, dot plots, violin plots, stacked bar plots, pseudotime plots).
Track-based data
bucket_urls(required): List of URLs pointing to S3 buckets containing BigWig or other supported files. See example and instruction Additional Information 1.genome(required): Reference genome build (e.g.hg38,mm10). All supported genome are Additional Information 2.ATAC_name(optional): List of custom display names for ATAC tracks, aligned withbucket_urls. Defaults toTrack1,Track2etc.max_height(optional): List of maximum display heights for tracks, aligned withbucket_urls. Defaults to autoscale.
General customization
Fields outside of individual studies apply globally to the entire GUANACO session:
title(optional): Title of the GUANACO instance.color(optional): List of custom HEX colors applied to clusters and tracks.
Example Config File
{
"Study1": {
"description": "PBMC single-cell RNA and ATAC integration study",
"sc_data": "pbmc.h5ad",
"markers": ["MS4A1", "LYZ"],
"genome": "hg38",
"bucket_urls": ["https://atac-bucket-1/"],
"ATAC_name": ["PBMC-ATAC"],
"max_height": [20]
},
"Study2": {
"description": "Mouse skin study",
"sc_data": "skin.h5mu"
},
"title": "GUANACO",
"color": ["#1f77b4", "#ff7f0e", "#2ca02c"]
}
Once the config file is set and the AnnData and BigWig files are prepared, GUANACO can be accessed locally or deployed on a server. The data will be displayed in the designated areas of the platform, as shown in the figures below.
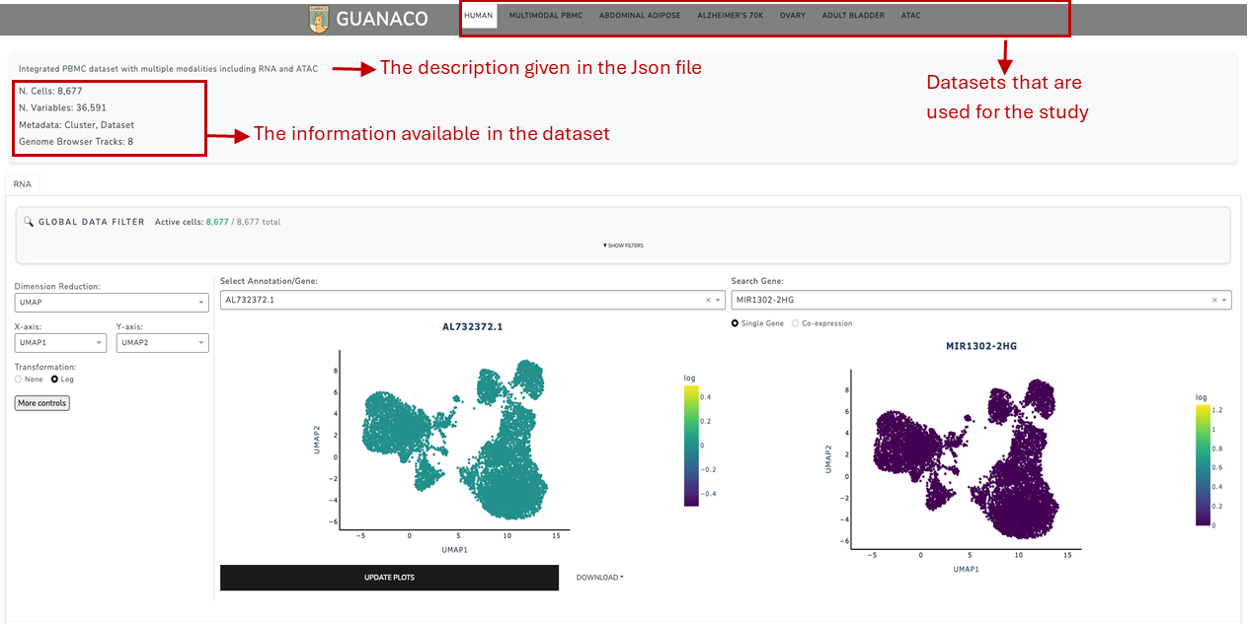
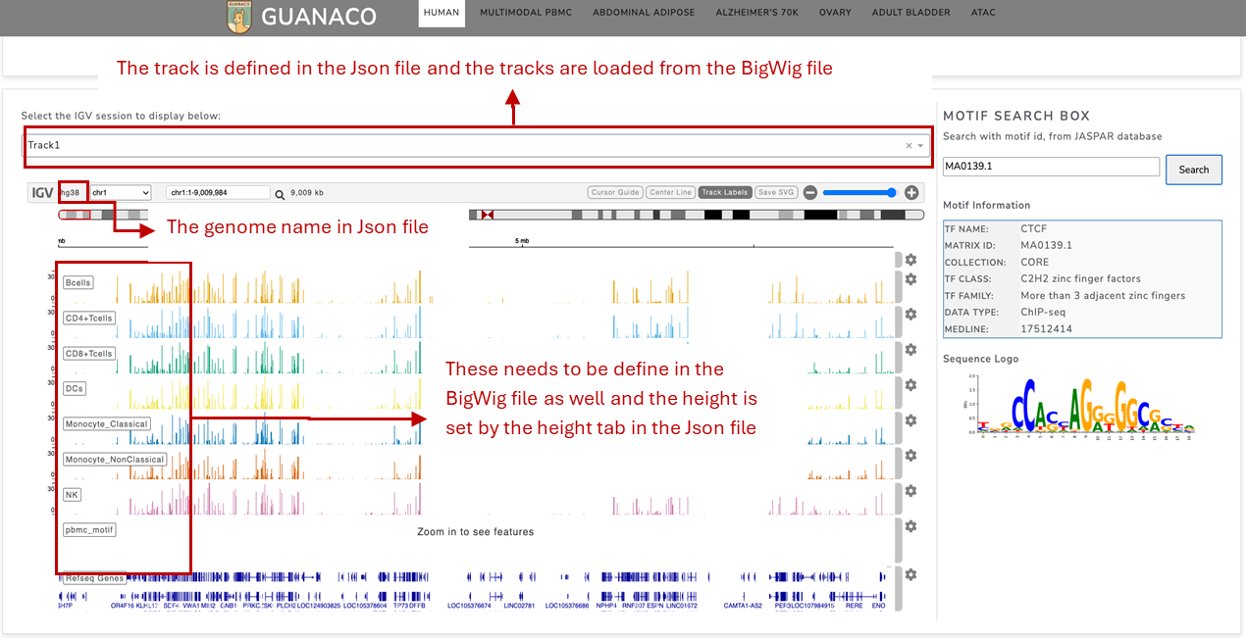
Additional Information
1. Track Data Configuration (Formats, URLs, Policies, CORS)
Users should upload BigWig or other supported files to a cloud storage service that provides public URLs. AWS S3 is recommended for simplicity and scalability.
Supported URL formats
AWS-style:
https://bucket.s3.region.amazonaws.comPath-style:
https://host/bucket[/prefix...]
Supported file formats
BigWig:
.bigwig,.bw(ATAC peaks)
If you need to convert an .h5ad or .h5mu file to BigWig, refer to the Jupyter notebook example provided with scCAMEL:
TACoWig_Template.ipynb
Interaction files:
.bedpeAnnotation tracks (motifs, SNPs, etc.):
.bed,.bigBed(.bb)You can generate motif tracks in BigBed format in two ways:
Using the JASPAR TFBS extraction tool: https://jaspar.elixir.no/tfbs_extraction/
Using our provided script: https://github.com/Systems-Immunometabolism-Lab/guanaco-viz/blob/main/motif_extraction
Bucket policy for public read access
Your S3 bucket must allow public read access so that GUANACO can fetch the files.
Use the following bucket policy as an example (replace name-of-your-bucket with your bucket name):
{
"Version": "2012-10-17",
"Statement": [
{
"Sid": "PublicListBucket",
"Effect": "Allow",
"Principal": "*",
"Action": "s3:ListBucket",
"Resource": "arn:aws:s3:::name-of-your-bucket"
},
{
"Sid": "PublicReadGetObject",
"Effect": "Allow",
"Principal": "*",
"Action": "s3:GetObject",
"Resource": "arn:aws:s3:::name-of-your-bucket/*"
}
]
}
CORS configuration
To enable cross-origin requests (web browser access), configure CORS on your bucket. Example configuration:
[
{
"AllowedHeaders": ["*"],
"AllowedMethods": ["GET", "HEAD"],
"AllowedOrigins": ["*"],
"ExposeHeaders": ["ETag"],
"MaxAgeSeconds": 3000
}
]
⚠️ Security note: For better security, replace "*"
in AllowedOrigins with a specific domain (e.g. "http://example.com") to restrict access.
2. Supported genome builds for track-based data
GUANACO supports the following reference genomes (UCSC 2bit format):
Human:
hg38,hg19,hg18Mouse:
mm39,mm10,mm9Rat:
rn6,rn5Zebrafish:
danRer11,danRer10Fruit fly:
dm6,dm3Worm (C. elegans):
ce11,ce10Yeast:
sacCer3Chicken:
galGal6Xenopus:
xenTro9Dog:
canFam3Cow:
bosTau9Pig:
susScr11Macaque:
rheMac10Each build corresponds to a UCSC-hosted 2bit file, e.g.:
https://hgdownload.cse.ucsc.edu/goldenPath/hg38/bigZips/hg38.2bit